RESEARCH
STUDYING ENHANCER AT ALL SCALES – Deciphering the molecular basis of human enhanceropathies
The primary goal of ENHPATHY is to provide early-stage researchers (ESRs) with a multidisciplinary training programme in which cutting-edge genomics and genome editing approaches are combined with in vitro and in vivo disease models to enable understanding the molecular basis of human enhanceropathies. Moreover, together with our private partners, we will explore the translation of our molecular findings into new diagnostic and therapeutic avenues for patients.
Enhancers encompass a diverse group of regulatory sequences, which utilize various mechanisms to control gene expression at the transcriptional and/or post-transcriptional level causing a broad range of functional effects. Not surprisingly, deregulation of enhancers by either genomic alterations (amplification, deletion, translocation, inversion, mutations) or epigenetic mechanisms is a major cause of gene deregulation in cancer and congenital diseases. Therefore, enhanceropathies, namely diseases caused by the alteration of enhancer activity, range from developmental (e.g. polydactyly) and autoimmune diseases (e.g. diabetes) to cancer. Consequently, human enhanceropathies are likely to display a broad variety of molecular mechanisms that can only be uncovered using highly multidisciplinary Systems Biology approaches. Chiefly, elucidating the molecular basis of human enhanceropathies has far reaching translational implications, especially considering the pandemic proportions that some of these disorders are attaining in recent years.
We have divided our research programme into three highly complementary and interconnected scientific work packages (WP) (Figure 1) to address the following key questions:
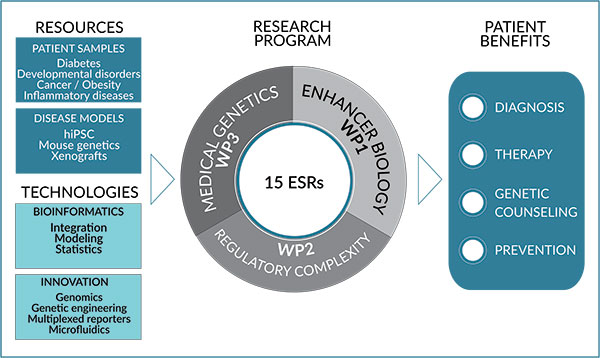
Figure 1 : A multidisciplinary research program linking enhancer biology and human diseases
WP1 – How do genetic and epigenetic features control enhancer activity?
By using combination of various (epi)genomic approaches and systematic reporter assays, we will study how transcription factor binding and genetic/epigenetic variation impact on enhancer activity.
ESR 1 project : Dissection of coordinated stress responses by novel promoter elements with enhancer activity (WP1)
We recently demonstrated that a subset of gene promoters, termed Epromoters, works “also” as bona fide enhancers and regulate distal gene expression. Subsequently, we found that Epromoters play a key role in the coordination of rapid gene induction in the stress response, in particular during inflammation. The ESR will study the functional role of Epromoters in different stress and inflammatory conditions, determine their impact in genome topology, and establish in vivo models to assess Epromoter variation in inflammatory diseases. This systematic approach will unravel the basic molecular mechanisms of this novel type of regulatory elements and shed the basis of their potential involvement in diseases.

Jing Wan (ESR1) individual web page
Supervisor : Salvatore Spicuglia
Coordinator & WP7 Leader
Institute Nationale pour la Santé et la Recherche Médicale (Inserm)
ESR 9 project : Analysis of the role of coregulators in the activity of cis-regulatory elements in response to inflammatory stimulation (WP1)
The project will address the mechanisms that control the function of cis-regulatory elements (enhancers and promoters) in basal conditions and in response to acute stimulation using the model of macrophages activated by inflammatory stimuli. In particular, the RNA Polymerase II recruited to both enhancers and promoters can generate a substantial amount of non-coding transcripts such as enhancer RNAs (eRNAs) and promoter-antisense RNAs. However, non-coding transcription efficiency at cis-regulatory elements is much lower compared to protein coding genes, suggesting the existence of mechanisms that attenuate RNA Pol II activity at these sites or conversely mechanisms that selectively enhance transcription within gene units. The work will entail the generation and computational analysis of multiple genomics data sets in normal cells or cells lacking a panel of novel candidate regulators.

Gaurav Mandana (ESR9) individual web page
Supervisor : Gioacchino Natoli
WP1 Leader
ESR 12 project : Regulatory network guiding stem cell interactions within the hematopoietic niche using single cell genomics (WP1)
Integrating data from ou in vitro model of the bone marrow niche with publicly available data, this project will reveal regulatory interactions between enhancers and their target genes that are triggered by the interaction of HSCs with different types of niche cells. Regulatory interactions will be derived from co-variation of regulatory elements (ATAC-Seq) and gene expression (RNA-Seq) across individuals. Specifically, we will investigate the effect of mesenchymal stromal cells (MSCs) and MSC-derived cell types (adipocytes and osteoblasts) on the regulatory network of HSCs. These comparisons are particularly interesting since the bone marrow niche becomes more adipocyte-dense during aging. At the same HSCs show a differentiation bias of HSCs from lymphoid to myeloid lineages upon ageing, a phenomenon that has been linked to the weakening of the adaptive immune system with old age. Integrating the regulatory network with SNPs associated with common (immune-system related) diseases, as obtained from genome-wide association studies, will reveal how the age-dependent compositional change in the bone marrow niche might contribute to disease phenotypes.

Maksim Kholmatov (ESR12) individual web page
Supervisor : Judith Zaugg
European Molecular Biology Laboratory
ESR 13 project : Functional annotation 10 human genomes to aid the identification of disease-causing non-coding sequence variants (WP1)
We have previously shown how our SuRE assay can be used to test non-coding sequence variants for their effect on promoter and enhancer activity. In this project we will establish a database with millions of non-coding sequence variants – all functionally annotated for their effect on enhancer and promoter activity in several disease-relevant cell types. We believe this will be an important resource for strategies aiming to link genotypes to phenotypes (e.g. GWAS and eQTL studies). The task of the student will be to develop effective methods for the integration, analysis and visualization of these large-scale datasets.

Vartika Bisht (ESR13) individual web page
Supervisor : Joris van Arensbergen
Legal Supervisor : SALVATORE SPICUGLIA
WP2 – How do enhancers function within complex regulatory landscapes?
We will study the mechanisms by which individual enhancers control distal gene regulation (Pol II recruitment and release, transcript maturation, priming, transcription robustness, genome topology) as well as the precise role of individual enhancers within clusters of enhancers.
ESR 5 project : Understanding the impact of gene regulatory variation on disease using variable chromatin modules (VCMs) (WP2)
In this project, we will leverage the transformative power of VCMs to unravel how genetic and regulatory variation guides phenotypic diversity. We predict that the most likely causal variants are those that associate with VCM activity, gene expression, and organismal phenotypes (here, extent of differentiation). We will map VCMs both at the single cell and bulk level and exploit their coordinated molecular nature to uncover the flow of regulatory information, from causal nucleotides over gene(s) to phenotype. As such, we aim to provide unprecedented insights into the molecular mechanisms driving phenotypic variation.

Olga Pushkareva (ESR5) individual web page
Supervisor : Bart Deplancke
Swiss Federal Institute of Technology Lausanne
ESR 7 project : Higher-order topological organization of super-enhancer hubs, Epromoters and TADs (WP2)
The project will characterize the functioning and impact of novel key topological factors on long-range regulatory and architectural chromatin interactions and hub formation, using state-of-the-art protein depletion, chromatin topology and microscopy technologies.

Anna Karina Felder (ESR7) individual web page
Supervisor : Wouter de Laat
Hubrecht Institute
ESR 8 project : Enhancer networks in the control of lineage determination of human mesenchymal stem cells (hMSCs) (WP2)
The project will investigate functional and mechanistic aspects of the enhancer networks regulating adipocyte differentiation of human mesenchymal stem cells. In unpublished work from our laboratory, we have shown that differentiation is driven by highly connected connected enhancer communities that appear to be preprogrammed already at the stem cell state. In this project we will determine the mechanisms by which these enhancer communities are activated during differentiation.

Anna Cetnarowska(ESR8) individual web page
Supervisor : Susanne Mandrup
WP2 Leader
ESR 10 project : The spatial organization of enhancers around promoter regions within chromatin contact domains for selected Human cell lines: structural regulatory landscape (WP2)
The objective of the research is to develop and test the concept of the structural landscape for regulatory elements around promoter regions for selected cell lines. We will propose novel biophysical method to construct probabilistic ensembles of three-dimensional conformations for chromatin contact domains (CCDs, sometimes described as TADs: topologically associating domains) at the whole genome scale. The computational method exploits results from two independent experimental sources: first the genomic-based interaction data from ChIA-PET together with epigenetic modifications and transcription factors binding sites occupancy (ChIP-seq), and secondly the mRNA expression profiles (RNA-seq) measured in the same cell lines. We will independently validate our findings and computational algorithms by extensive analysis of GWAS in relevant regulatory elements.

Abhishek Agarwal (ESR10) individual web page
Supervisor : Dariusz Plewczynski
University of Warsaw
ESR 14 project : Microfluidic systems for encapsulation of molecular complexes (WP2)
The ESR project will develop into two main axes. We will support the other teams within ENHPATHY working with microfluidics (EPFL, KNAW and KTH) in order to provide them with our flow control system. In particular we will partner with Dr. Deplancke’s laboratory to support them in the development of their system. On the other hand, the ESR will lead, in collaboration with Dr. Sahlén’s group, the development of an automated microfluidic system for double encapsulation droplet production.

Robert Baber (ESR14) individual web page
Supervisor : Julia Sepulveda
Legal Supervisor : PELIN SAHLÉN
WP3 – How does enhancer deregulation impact human diseases?
We will identify disease-causing enhancer alterations, develop approaches to discern pathogenic enhancer variants from neutral variants, study their pathological consequences, and set the ground for the development of new enhancer-based diagnostic and therapeutic tools.
ESR 2 project : Modeling gene-environmental interactions implicated in the etiology of human neurocristopathies (WP3)
Using human neural crest cells and the TFAP2A locus as a medically relevant models, our work will evaluate whether genetic variants within enhancers might interact with teratogenic factors as part of the etiology of human congenital abnormalities. Thus, by considering enhancers as hubs of gene-environmental interactions, our results can illuminate a novel and broadly relevant etiological paradigm for human disease.

Sarah Malika Robert (ESR2) individual web page
Supervisor : Alvaro Rada-Iglesia
Vice-coordinator & WP4 Leader
University of Cantabria
ESR 3 project : Genetic and epigenetic changes in hematopoietic stem cells (HSCs) during chronic inflammation (WP3)
This project will determine the molecular changes in enhancers and promoters that chronic inflammation induces in hematopoietic stem cells. Together, the results of this project will uncover the mechanisms regulating hematopoietic stem cell maintenance and fate in the context of chronic inflammation, and lay the path to identify new therapeutic strategies.

Sladana Kosanovic (ESR3) individual web page
Supervisor : Meritxell Alberich-Jorda
WP5 Leader
Institute of Molecular Genetics, Prague, Czech Republic
ESR 6 project : Computational approaches to discover noncoding mutations underlying human disease (WP3)
Human genetics is rapidly moving from the analysis of protein-coding sequences to whole genome sequences. Our ability to discern non-protein coding mutations that cause disease from a vast excess of non-functional mutations is still limited. This PhD project will focus on an ERC-funded large-scale enhancer mutation screen in patients with monogenic diabetes. It will develop computational approaches to inform on the pathogenicity and function of enhancer mutations, using data from ongoing experimental and patient screens.
Applicants with diverse educational backgrounds are welcome, but a computing background is important, including basic knowledge of a programming language (e.g Python, R). Expertise in statistical analysis is also highly relevant.
Supervisor : Jorge Ferrer
WP3 Leader
ESR 11 project : Functional understanding of synergistic effects of enhancer elements at the onset of atherosclerosis (WP3)
The aim of the project is to establish a method that can resolve multiple promoter-anchored interactions at single cell resolution. The method will utilise droplet technology and chromatin immunoprecipitation to identify multiple promoters and enhancers contacts. Such data will help elucidate to which degree enhancers cooperate in regulating expression of their target genes, and contribute to our understanding of the role of enhancer redundancy in complex diseases context. We will apply the methodology to aortic endothelial and smooth muscle cells, liver cells and activated macrophages to study the extent of redundancy in regulatory networks concerning cardiovascular risk variants.

Artemy Zhigulev (ESR11) individual web page
Supervisor : Pelin Sahlén
Royal Institute of Technology
ESR 15 project : Uncovering drug-adaptive epigenetic response in Acute Myeloid Leukemia (AML) (WP3)
This project will uncover drug-responsive regulatory elements and related molecular mechanisms that may have an impact on AML therapeutic outcomes and drug resistance. These results can pave the way to strategies with improved therapeutic efficacy by targeting of enhancers or their targets with inhibitors that can attenuate the adaptive transcription to genes related to drug resistance and proliferation.

Francesco Leonetti (ESR15) individual web page
Supervisor : Mileidys Perez
Legal Supervisor : SALVATORE SPICUGLIA
ESR 4 project : The impact of genetic variants on regulatory activities (WP3)
This project aims to explore the impact of regulatory genetic variation. In particular, the major aims are to establish accurate and interpretable machine learning approaches to learn how DNA sequence influences enhancer and promoter activities and to use derived models to interpret the determinants of regulatory function and the impact of regulatory genetic variation on their activities in health and disease. To this end, part of the project will focus on in vivo (CAGE, ATAC-seq, Hi-C) and in vitro (STARR-seq, SURE-seq) data generated in our lab and within the ENHPATHY network across a genotyped panel of lymphoblastoid cell lines, displaying large individual regulatory variation. Inferred models will further our understanding of the determinants of regulatory function and improve genetic variant interpretation, which will be useful for a large scientific and medical community, ultimately leading to insights into why and when regulatory genetic variation may cause phenotypical or pathological changes.

Natsuda Navamajiti (ESR4) individual web page
Supervisor : Robin Andersson
University of Copenhagen
QUESTION ABOUT ENHPATHY?


![]() This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 860002. The information contained in this website reflects only the authors’ view. REA and EC are not responsible for any use that may be made of this information.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 860002. The information contained in this website reflects only the authors’ view. REA and EC are not responsible for any use that may be made of this information.
This site uses cookies. By continuing to browse the site, you are agreeing to our use of cookies.
Accept settingsHide notification onlyParamètresWe may request cookies to be set on your device. We use cookies to let us know when you visit our websites, how you interact with us, to enrich your user experience, and to customize your relationship with our website.
Click on the different category headings to find out more. You can also change some of your preferences. Note that blocking some types of cookies may impact your experience on our websites and the services we are able to offer.
These cookies are strictly necessary to provide you with services available through our website and to use some of its features.
Because these cookies are strictly necessary to deliver the website, refuseing them will have impact how our site functions. You always can block or delete cookies by changing your browser settings and force blocking all cookies on this website. But this will always prompt you to accept/refuse cookies when revisiting our site.
We fully respect if you want to refuse cookies but to avoid asking you again and again kindly allow us to store a cookie for that. You are free to opt out any time or opt in for other cookies to get a better experience. If you refuse cookies we will remove all set cookies in our domain.
We provide you with a list of stored cookies on your computer in our domain so you can check what we stored. Due to security reasons we are not able to show or modify cookies from other domains. You can check these in your browser security settings.
We also use different external services like Google Webfonts, Google Maps, and external Video providers. Since these providers may collect personal data like your IP address we allow you to block them here. Please be aware that this might heavily reduce the functionality and appearance of our site. Changes will take effect once you reload the page.
Google Webfont Settings:
Google Map Settings:
Google reCaptcha Settings:
Vimeo and Youtube video embeds:
You can read about our cookies and privacy settings in detail on our Privacy Policy Page.
Legal Notice